CoVe-tracker is an interactive database that provides the evolution of SARS-CoV-2 (PANGO) lineages (lineage) and the concomitant variants and mutations.
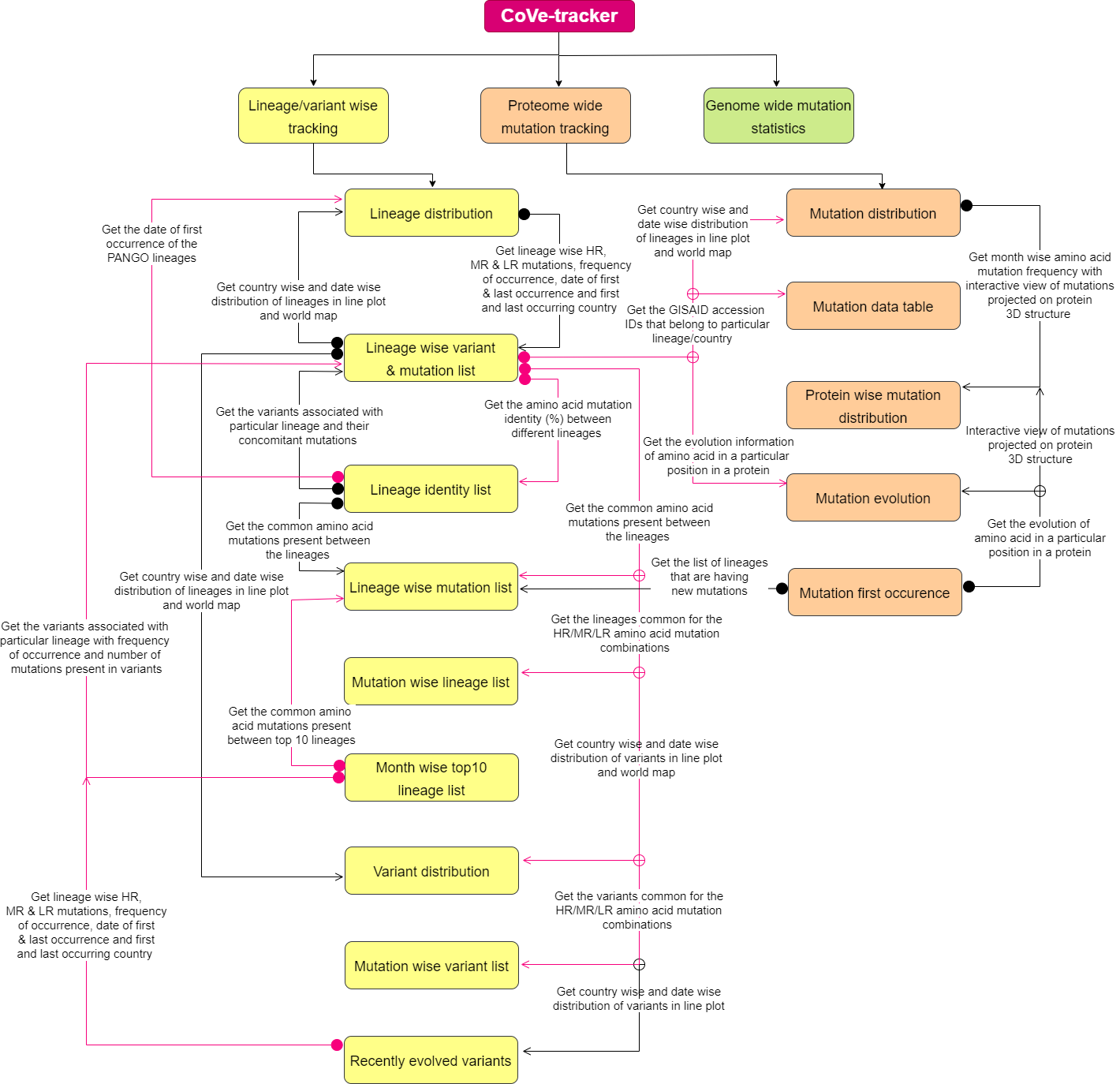
CoVe-tracker comes with the following three main menus: Lineage/variant wise tracking, Proteome wide mutation tracking, and Genome wide mutation statistics.
The “Lineage/variant wise tracking” menu bar has nine submenus, using which the user can track country-wise and month-wise distribution of any lineage and variant and obtain lineage-wise variant and mutation lists, lineage identity lists, lineage-wise mutation lists, mutation-wise lineage and variant lists, month-wise top 10 lineage list, and the variant evolution trend during the past 2 months.
Under the “Proteome wide mutation tracking” option, the following submenus are displayed: mutation tracking, mutation data table, protein-wise mutation distribution, mutation evolution, and the first occurrence of the mutation.
The “Genome wide mutation statistics” main menu provides a scatter plot of position-wise nucleotide mutation frequency and a mutation data table. The lineage, variant, amino acid mutation, and genomic mutation information of SARS-CoV-2 can be accessed through the respective menu bar. Thus, these options in CoVe-tracker provide the country-wise statistics of all the existing variants, lineages, and mutations. The month-wise statistics of the lineages and variants provide their emergence and dissemination during the selected timeline. Notably, CoVe-tracker also provides high, moderate, and low recurring (LR) amino acid mutations. CoVe-tracker further lists the month-wise top 10 lineages and the concomitant variants to track the evolution of a new lineage/variant.
CoVe-tracker thus provides the evolution of SARS-CoV-2 lineages, variants, mutations and their dissemination. Since CoVe-tracker is updated every month using the SARS-CoV-2 sequences from GISAID (Dec 2019 to Dec 31, 2024), it provides up to date information on the viral evolution.
Statistics
Total number of sequences considered: 3344714
Total number of amino acid mutations: 51479
Highly recurring (HR) mutations: 52
Moderately recurring (MR) mutations: 117
Low recurring (LR) mutations: 36407
Total number of nucleotide mutations: 118361
Total number of lineages: 2584
Total number of variants: 1410580
 Last upadte: 03 Jul, 2025 Data considered from GISAID: till 31 May, 2025
Last upadte: 03 Jul, 2025 Data considered from GISAID: till 31 May, 2025