Overview
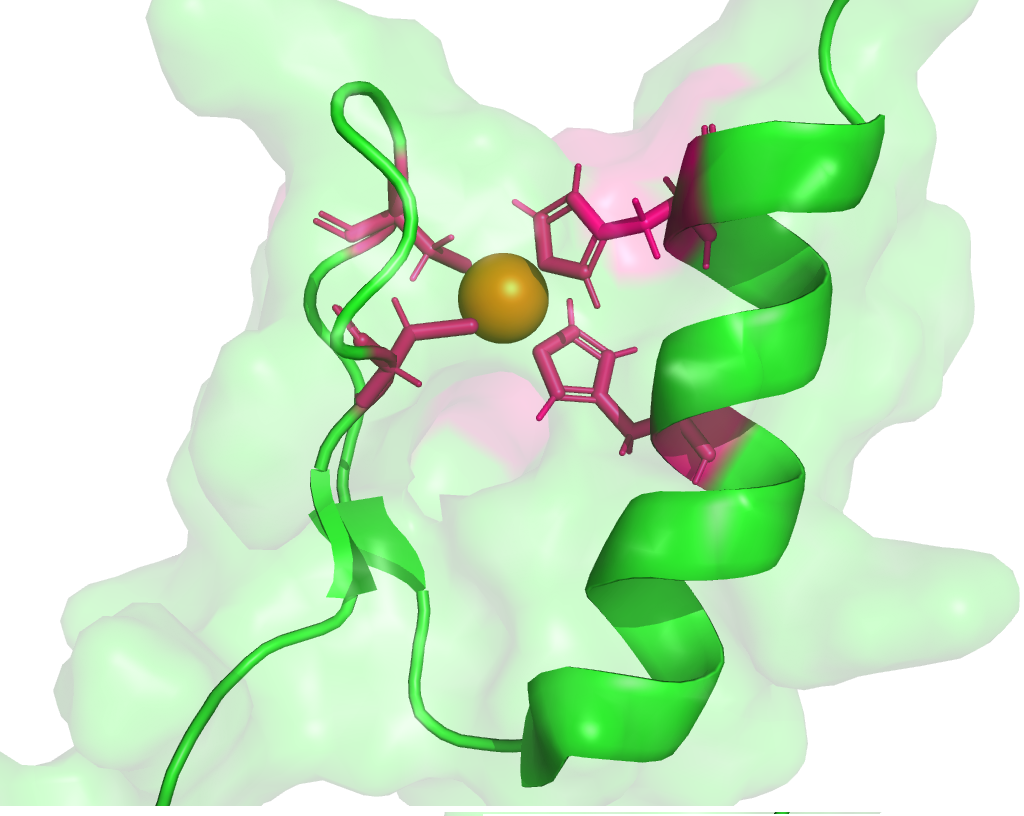
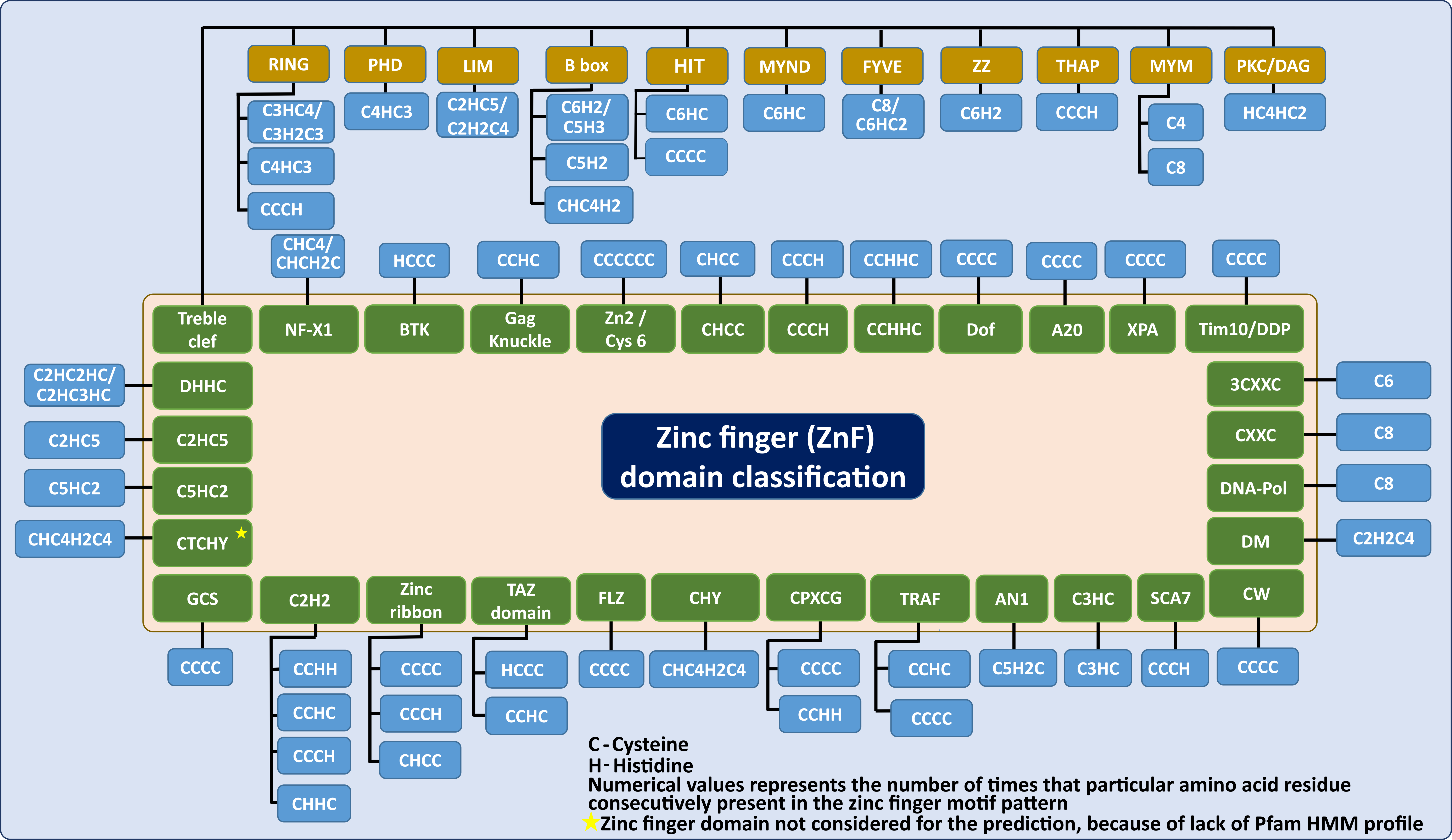
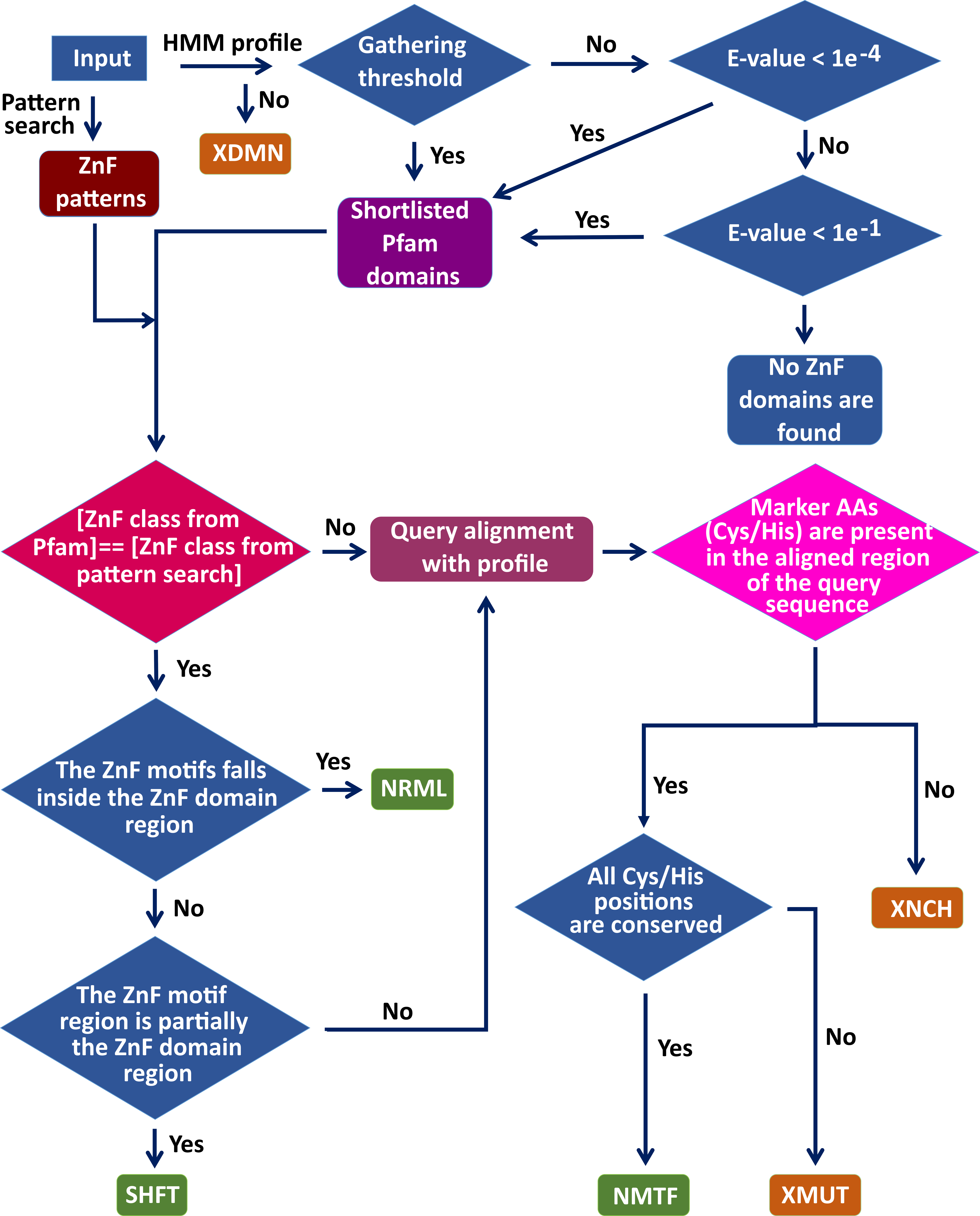
Zinc finger (ZnF) is a class of small protein domains, wherein, Zn(II) is the inorganic co-factor that forms a tetrahedral geometry with the cysteine and/or histidine. It plays an important role in protein catalytic activity, stability and folding. Nonetheless, predicting the ZnF motif from the sequence is quite challenging. To this end, 71 unique ZnF motif patterns have been collected through the literature survey and used in the prediction of 31 different classes of ZnF domains with diverse functions. Since the short length of the ZnF motif patterns leads to inaccurate prediction (false positives), 288 Pfam (Protein families) HMM profiles corresponding to 6 ZnF Pfam clans (215 HMM profiles) and 73 HMM profiles from other undefined clans (but, belong to ZnF domains) have been used in the prediction. ZnF-Prot can predict the presence of ZnF motif(s) from a protein(s)/proteome sequence.
Click the below buttons to go respective pages
Video tutorial
The systematic zinc finger domain classification was carried out based on the references given below. User can click the link that is given below for detailed information.
Zinc-finger proteins in health and disease Metal binding properties, stability and reactivity of zinc fingers Caught with One's Zinc Fingers in the Genome Integrity Cookie Jar Structural classification of zinc fingers: SURVEY AND SUMMARY