Tutorial
User can click the below given button to predict the ZnF motif present in example query sequence (FASTA sequence from PDB ID: 1VA1)
Znf motif prediction - step wise explaination
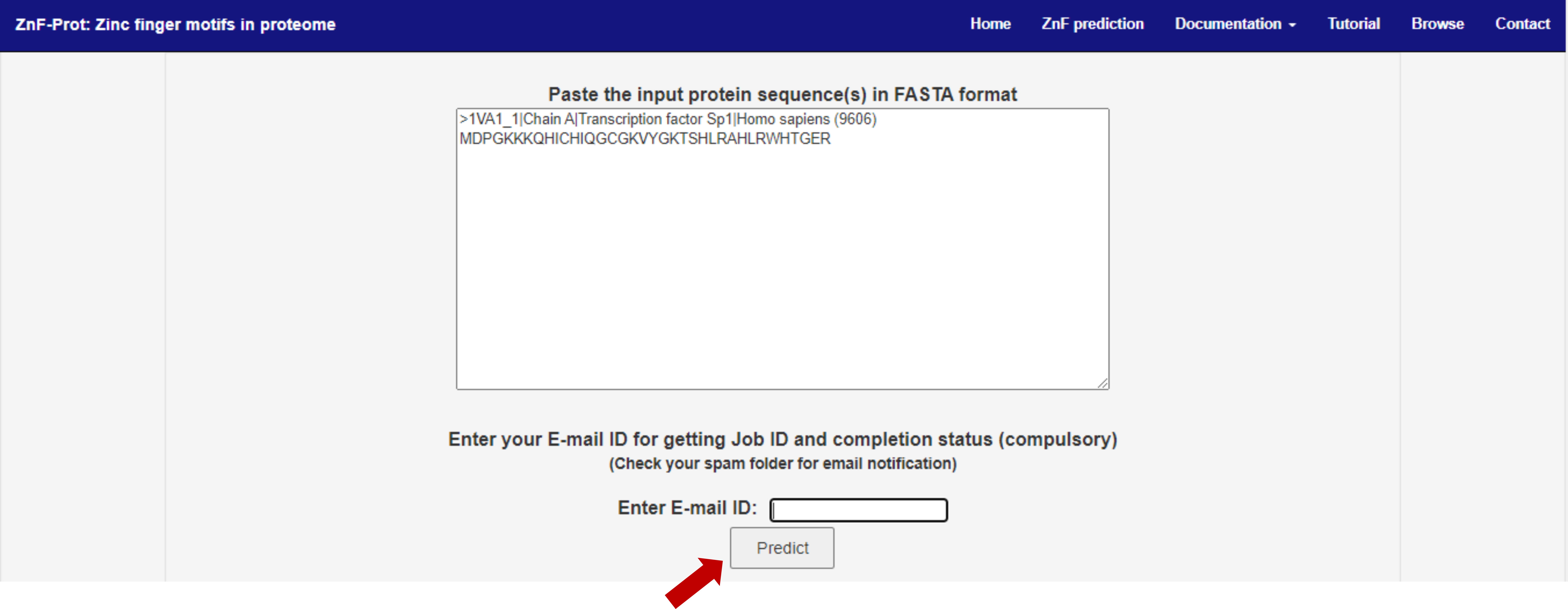
1. User can paste the input query sequence (protein) in FASTA format and click the predict button (indicated with red arrow mark in the figure)
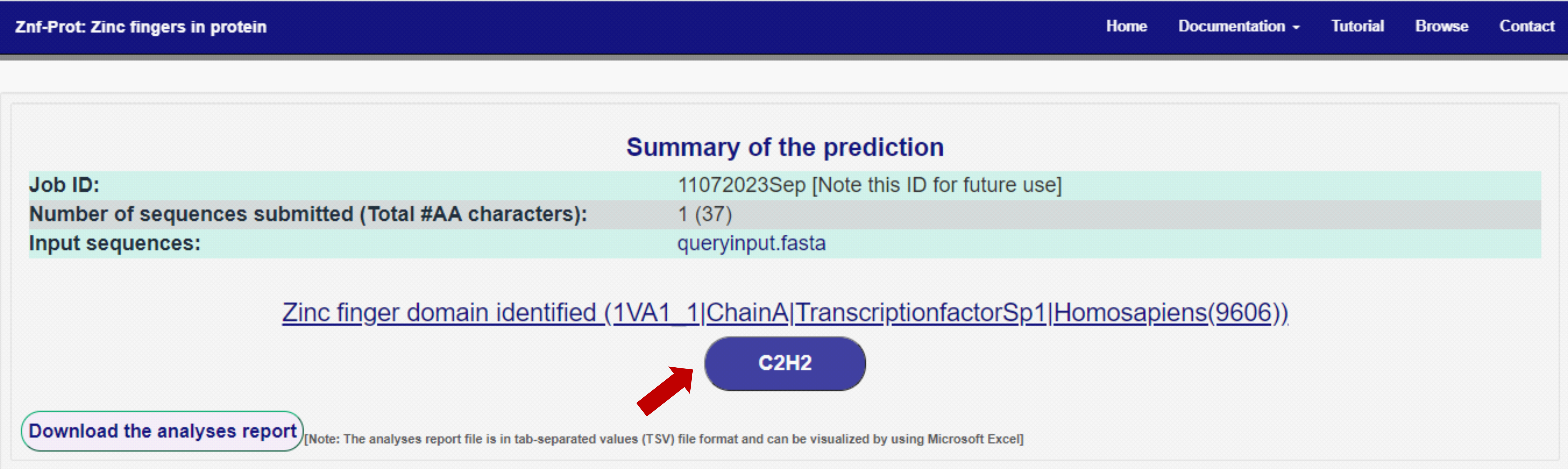
2. Summary of the prediction: It will display sequence-wise ZnF motif prediction results. For example, if users submit more input sequences, it will show sequence-wise prediction results. Since the submitted input sequence was single, the prediction result was displayed for a single sequence in the displayed test case. The result is displayed in the blue-colored box (C2H2)(indicated with a red arrow mark in the figure). Note: The user can note the Job Id for further use. Users can submit the Job Id on the "Browse" page to view the results back.
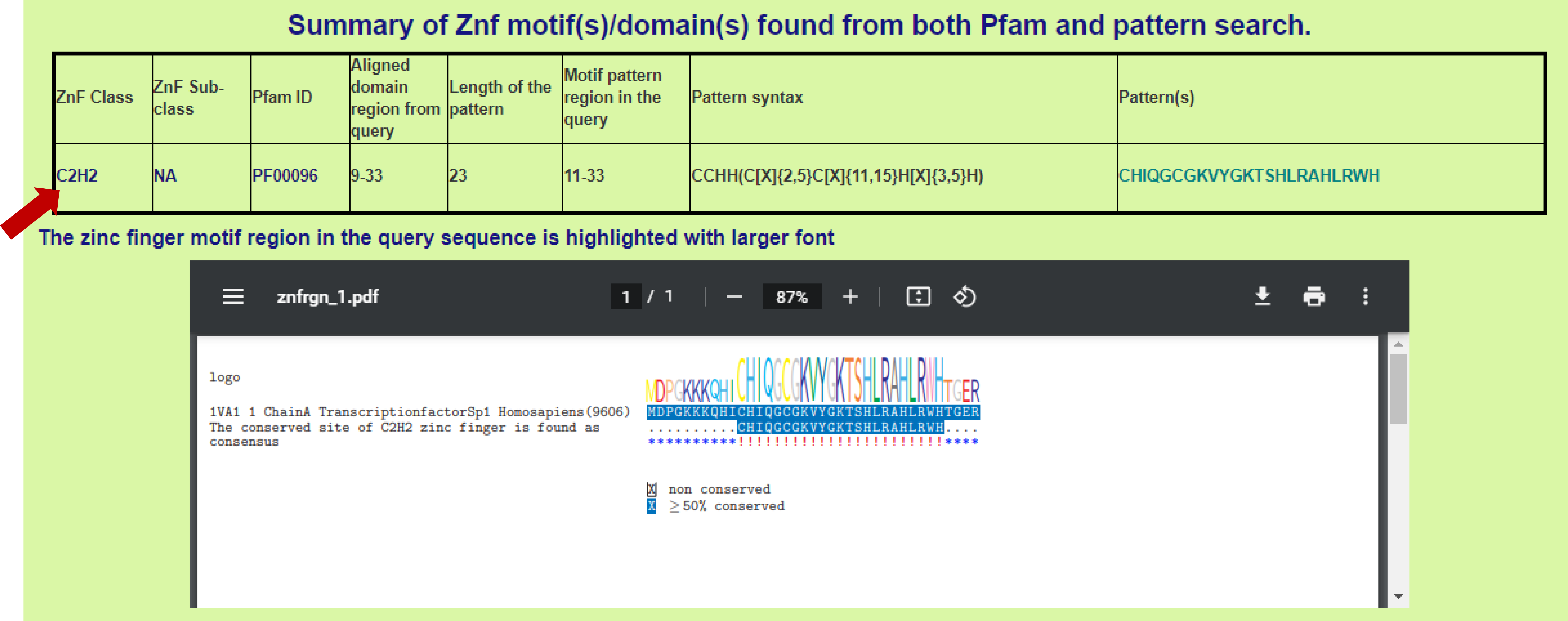
3. Sequence-wise ZnF prediction - Results in detail (Summary of ZnF motifs found from both ZnF pattern search and Pfam HMM profile search approach): The detailed information of the ZnF prediction will be displayed here. The result will be displayed in 8 columns. 1st column displays the ZnF class identified from the input query sequence (indicated with red arrow mark in the figure), 2nd column displays the ZnF sub-class (NA indicates not available), 3rd column displays the Pfam accession ID corresponding to the domain present in query sequence, 4th column displays the aligned Pfam domain region from the query, 5th column displays the length of identified ZnF motif pattern, 6th column displays region of the ZnF motif pattern in the query sequence (starting to ending position), 7th column displays the identified ZnF motif pattern syntax (C[X]{2,5}C[X]{11,12}H[X]{3,5}H). C and H represent cysteine and histidine amino acid residues that coordinating with Zn ion, [X] represents any amino acids, numerical value represents the number of amino acid residues and [X]{2,5} represents 2 to 5) amino acid residues in the pattern and 8th column represents the ZnF domain sequence region present in input sequence. Pictorial representation highlighting the ZnF domain region in the input query sequence.