Help:
What are the various ID types?
1. AlphaFold Accessions: These accessions can be mapped
directly
from UniProtKB Accessions to download structural coordinate files from AlphaFold Database.
2. UniProt Accessions: These are 6 or 10 alphanumerical
character
long stable identifiers for UniProtKB entries.
They can be directly searched AlphaFold Structure Database. Click here to know
more.
3. RefSeq Accessions: The NCBI Reference Sequence (RefSeq)
project
provides non-redundant curated data of sequence records and
related information for numerous organisms, and provides a baseline for medical, functional, and
comparative studies. A RefSeq record can be identified by its distinct
accession number format that begins with two characters followed by an underscore (e.g., WP_). The
prefixes of these accessions are of various types. Click here to
know
more.
4. Locus Tags: Locus_tags are identifiers that are
systematically
applied to every protein coding and non-coding gene in a genome. Their prefixes are unique to every
organism and
there is only one locus_tag associated with one gene. The locus_tag prefix can contain only
alpha-numeric characters and it must be at least 3 characters long. Click here to
know
more.
5. Old Locus Tags: The old locus tags were used prior to the
Prokaryotic Genome Annotation Pipeline (PGAP) re-annotation and the locus tags are the newer
annotation.
On the current RefSeq bacterial genome record, the /old_locus_tag qualifier is still annotated. Click
here
to know more.
6. TaxIDs: Each NCBI Taxonomy entry or TaxNode is assigned a
public
stable, unique numerical identifier-the taxonomy identifier (TaxId)-which is shared by all names for
a
specific TaxNode.
Click here to know
more.
Sample Inputs and Outputs:
If the input is AlphaFold Acessions or UniProt Accessions, the respective AlphaFold prediction will be downloaded. However, it is possible that some UniProt accessions will not have AlphaFold predicted structures, so no files will be downloaded for such cases.
| AlphaFold IDs |
|---|
| AF-Q9ULZ0-F1 |
| AF-Q8R143-F1 |
| AF-Q9LU47-F1 |
| AF-P05453-F1 |
| AF-Q7WTR3-F1 |
or
| UniProt Accessions |
|---|
| Q9ULZ0 |
| Q8R143 |
| Q9LU47 |
| P05453 |
| Q7WTR3 |
| Structure files downloaded |
|---|
| AF-Q9ULZ0-F1.pdb, AF-Q9ULZ0-F1.cif, AF-Q9ULZ0-F1_error.json |
| AF-Q8R143-F1.pdb, AF-Q8R143-F1.cif, AF-Q8R143-F1_error.json |
| AF-Q9LU47-F1.pdb, AF-Q9LU47-F1.cif, AF-Q9LU47-F1_error.json |
| AF-P05453-F1.pdb, AF-P05453-F1.cif, AF-P05453-F1_error.json |
| AF-Q7WTR3-F1.pdb, AF-Q7WTR3-F1.cif, AF-Q7WTR3-F1_error.json |
For RefSeq identifiers as the input, they could map either to single or multiple species such as example 1 and 3 in the table below. In cases when no structure file is downloaded, the reason could be that there is no corresponding UniProt Accession (example 2) or no predicted structure (example 4).
| RefSeq Protein IDs |
|---|
| NP_666037.1 |
| WP_296409975.1 |
| WP_004157415.1 |
| NP_056986.2 |
| Mapped UniProt Accession | Structure files downloaded |
|---|---|
| Q8R143 | AF-Q8R143-F1.pdb, AF-Q8R143-F1.cif, AF-Q8R143-F1_error.json |
| Not found | No files downloaded |
| A0A831EQJ9 Q7WTR3 D4I2C8 |
The files corresponding to all 3 UniProt accessions will be downloaded |
| O95071 | No predicted structures for this accession are available on AlphaFoldDB - No files downloaded |
For old locus tags as the input, they are mapped to the corresponding UniProt accession. In cases when no structure file is downloaded, the reason could be that there is no corresponding UniProt Accession (example 2, 4) or no predicted structure (example 3).
| Old Locus Tags |
|---|
| GVO57_04600 |
| MXAN_1028 |
| L3078_27850 |
| CL52_18575 |
| LN051_09145 |
| Organism Name | Locus Tag | Old locus Tag | UniProt Accession | Files Downloaded |
|---|---|---|---|---|
| Sphingomonas changnyeongensis | GVO57_RS04600 | GVO57_04600 | A0A7Z2NV19 | AF-A0A7Z2NV19-F1.pdb, AF-A0A7Z2NV19-F1.cif, AF-A0A7Z2NV19-F1.bcif, AF-A0A7Z2NV19-F1_error.json |
| Myxococcus xanthus DK1622 | MXAN_RS04940 | MXAN_1028 | Q1DDI4 | AF-Q1DDI4-F1.pdb, AF-Q1DDI4-F1.cif, AF-Q1DDI4-F1.bcif, AF-Q1DDI4-F1_error.json |
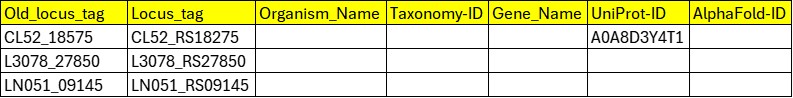
| Streptomyces deccanensis | L3078_RS27850 | L3078_27850 | Not found | No files downloaded |
| Stutzerimonas balearica DSM 6083 | CL52_RS18275 | CL52_18575 | A0A8D3Y4T1 | No predicted structures for this accession are available on AlphaFoldDB - No files downloaded |
| Staphylococcus ratti strain CCM 9025 | LN051_RS09145 | LN051_09145 | Not found | No files downloaded |
For locus tags as the input, they are first mapped to old locus tags and these old locus tags are then mapped to UniProt accessions. This is done because the old locus tags have a one-to-one association with UniProt accessions. No structure file is downloaded, when a corresponding UniProt accession (example 2) or predicted structure (example 4) is not found.
| Locus Tags |
|---|
| GVO57_RS04600 |
| MXAN_RS04940 |
| L3078_RS27850 |
| CL52_RS18275 |
| J0917_RS17640 |
| Organism Name | Locus Tag | Old locus Tag | UniProt Accession | Files Downloaded |
|---|---|---|---|---|
| Sphingomonas changnyeongensis | GVO57_RS04600 | GVO57_04600 | A0A7Z2NV19 | AF-A0A7Z2NV19-F1.pdb, AF-A0A7Z2NV19-F1.cif, AF-A0A7Z2NV19-F1.bcif, AF-A0A7Z2NV19-F1_error.json |
| Myxococcus xanthus DK1622 | MXAN_RS04940 | MXAN_1028 | Q1DDI4 | AF-Q1DDI4-F1.pdb, AF-Q1DDI4-F1.cif, AF-Q1DDI4-F1.bcif, AF-Q1DDI4-F1_error.json |
| Streptomyces deccanensis | L3078_RS27850 | L3078_27850 | Not found | No files downloaded |
| Stutzerimonas balearica DSM 6083 | CL52_RS18275 | A0A8D3Y4T1 | A0A8D3Y4T1 | No predicted structures for this accession are available on AlphaFoldDB - No files downloaded |
| Streptomyces nodosus | J0917_RS17640 | Not found | Not found | No files downloaded |
For NCBI TaxIDs as input, the structure files will be downloaded for all available AlphaFold predictions of proteins in that organism.
| Taxonomy IDs | Organism Name |
|---|---|
| 387662 | Candidatus Carsonella ruddii PV |
| Mapped UniProt Accessions for all proteins | Structure Files downloaded |
|---|---|
| Download mapped UniProt Accessions and details of all proteins in organism | Every protein whose structure files are available on AlphaFoldDB - will be downloaded |
Output files:
Apart from the user's choice of structure files, two other files can be found in downloaded zip file:
- The ID_mapping.csv file: This file provides the metadata associated with the input accessions. We
provide this to ensure that the user knows which AlphaFold accession corresponds to which input
accession because the downloaded structure files are named according to their AlphaFold accession.

- Missing_IDs.txt file: This file shows which of the user input acessions did not have a
corresponding UniProt/AlphaFold accession due to which their structure file could not be downloaded. It
has the same column headers as the ID_mapping.csv file.
Common errors and solutions:
No files downloaded?
1) Check your query type: You may have selected the wrong query type for your input accessions. We have provided some default examples for each accession format. Compare your query list with the examples to select the correct query type.
2) Input format: Make sure the input accessesions are new line separated and NOT by , or | or tab (\t) or space etc.
3) Submitting >5000 accessions: For large jobs of greater than 5000 input accessions, please use the API and not the webserver.
4) Download link expiry: Your zip files will be available to download only for two days. The files are deleted from our server immediately after you download them. Make sure to submit your e-mail ID for the larger jobs.
5) UniProt or AlphaFold accession doesn't exist: There's a possibility that the corresponding UniProt or AlphaFold accession for your input doesn't exist and in such cases no files will get downloaded. We have provided some examples such as WP_296409975.1, L3078_RS27850 and CL52_RS18275 in the sample inputs setion above. But still you may independenty verify your specific case on the corresponding database and in case you find your required prediction, reach out to us via the feedback form available on the Team Page
One to many accession correspondence
There is a probability that your input accession maps to more than one accession of another type, leading to multiple extra stucture prediction files getting downloaded. You may refer to the ID_mapping.csv file to pick your required structure based on the organism name, gene name and other important details provided there.

