About the tool:
Today there is an avalanche of predicted structures available on the AlphaFold Structure Database, which has
brought a huge
paradigm shift in the analysis and interpretation of protein functions. The access to the structure
files in
PDB and mmCIF formats makes
it easy for structural biologists and bioinformaticians to perform analyses. AlphaFold DB provides an
option
for 48 organisms’ bulk structure downloads
(model organisms and global health proteome) and otherwise very recently, the bulk download has been
enabled
for up to 100 structures. However, there is no direct way to selectively
download all the structures of interest at once. Also, several bioinformaticians work with sequence data
from NCBI and have access to various
protein
sequence identifiers but
cannot extract structures based on those sequence identifiers.
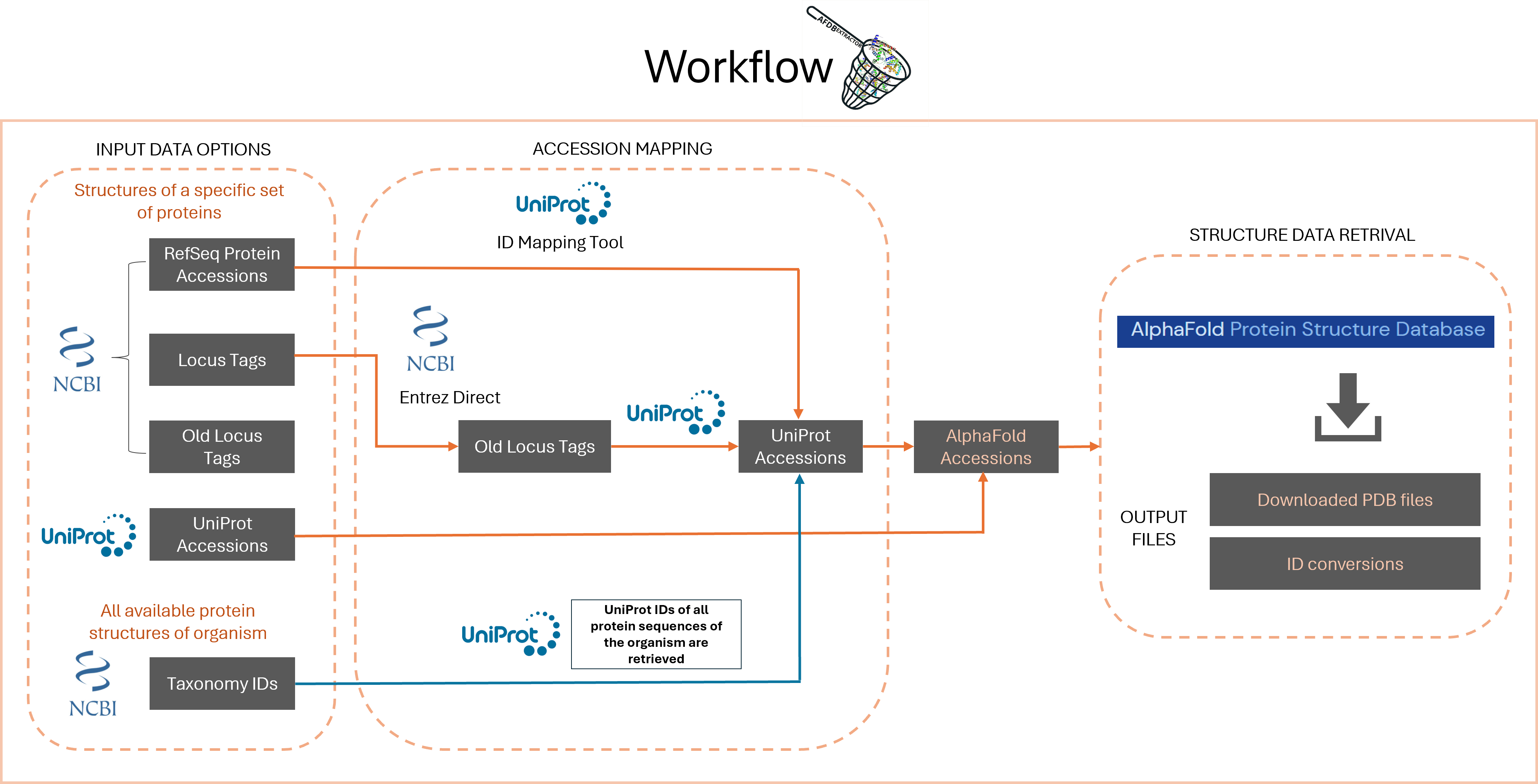
To bridge these gaps, we have
developed AlphaFoldDBExtractor which is a tool that lets users
download all available predicted structures from AlphaFoldDB for their dataset of interest with just a
list
of taxonomy/protein accessions.
Operating system(s): Platform independent.
Programming language: Shell scripting, Javascript, HTML, CSS
Other requirements: Firefox v109 or higher, Chrome v109 or higher
License: MIT
Any restrictions to use by non-academics: Not Applicable
This tool makes use of the AlphaFold Protein
Structure
Database, developed by
DeepMind and
EMBL–EBI, and released
under an
open-access policy. For details, see:
- Jumper, J. et al. (2021), Nature — Highly accurate protein structure prediction with AlphaFold .
- Varadi, M. et al. (2024), Nucleic Acids Research — AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences .
Input:
| List of specific proteins: |
|---|
| AlphaFold IDs |
| Uniprot IDs |
| Locus tags |
| Old locus tags |
| RefSeq Protein IDs |
| All proteins of organims: |
|---|
| NCBI TaxID |
Workflow: