SSP: Salmonella species serotyping web-GUI
Salmonella is a Gram-negative, facultatively anaerobic, rod-shaped (bacillus) bacteria of Enterobacteriaceae. They are non-spore-forming, predominantly motile enterobacteria with peritrichous flagella (all around the cell body, allowing them to move).
Salmonella is listed by the World Health Organization (WHO) as a “High Priority Pathogen” (priority 2), and urgently needs new antibiotics as they pose a significant threat to human health.
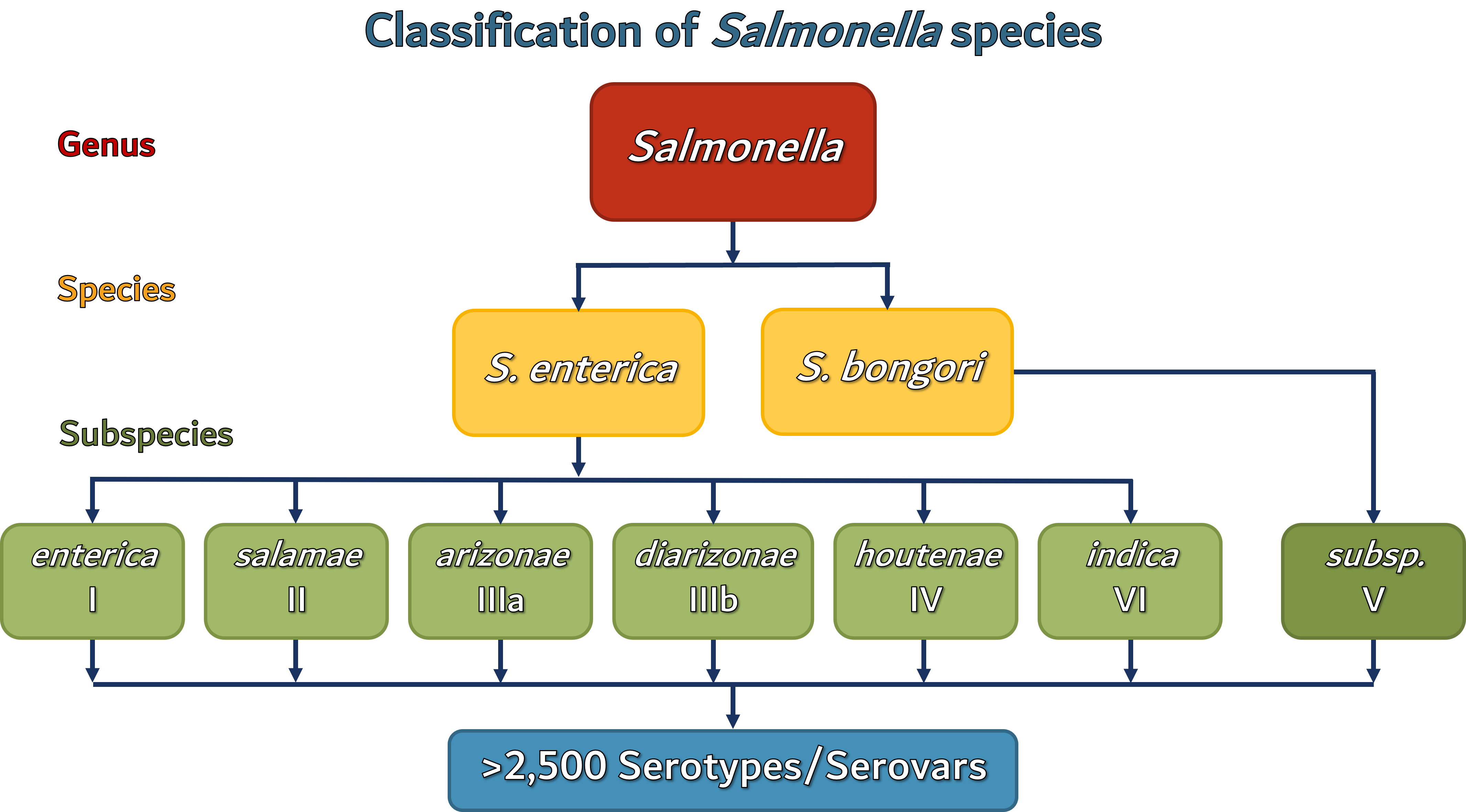
The genus Salmonella comprises two major species that include S. enterica and S. bongori. The following is the most recent classification used by the Center for Disease Control and Prevention (CDC):
The Kauffmann-White-Le Minor (KW) scheme is a system that is used to classify Salmonella into various serotypes and is based on the somatic (O) and flagellar (H) antigens present on its surface. The O-antigen type is determined based on the nature of the oligosaccharides associated with the lipopolysaccharide (LPS), and the H-antigen is determined based on the flagellar proteins. The H-antigens of Salmonella exhibit phase variation, and hence different H-antigens may be expressed.
There are more than 2500 species that fall under the Salmonella genus which are classified based on the combination of O- and H- (viz., H1 and H2) antigens using one species-one serotype principle.
The O-antigen determines the serogroup while the H-antigen completes the definition of serotype or serovar of a Salmonella isolate. The serotype is represented as follows:





