AagingPEPred: Anti-aging Peptide Prediction Server
Welcome to AagingPEPred
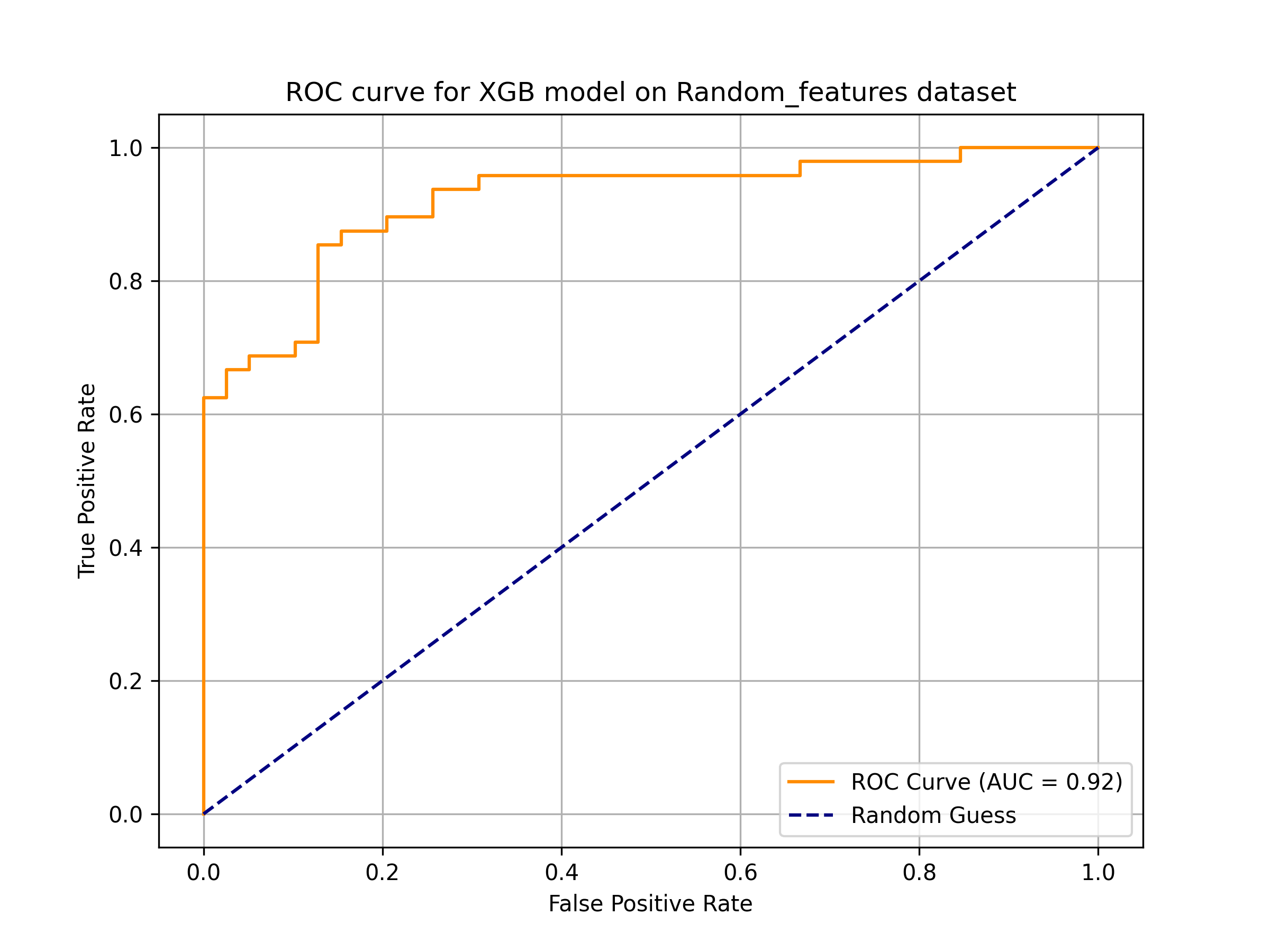
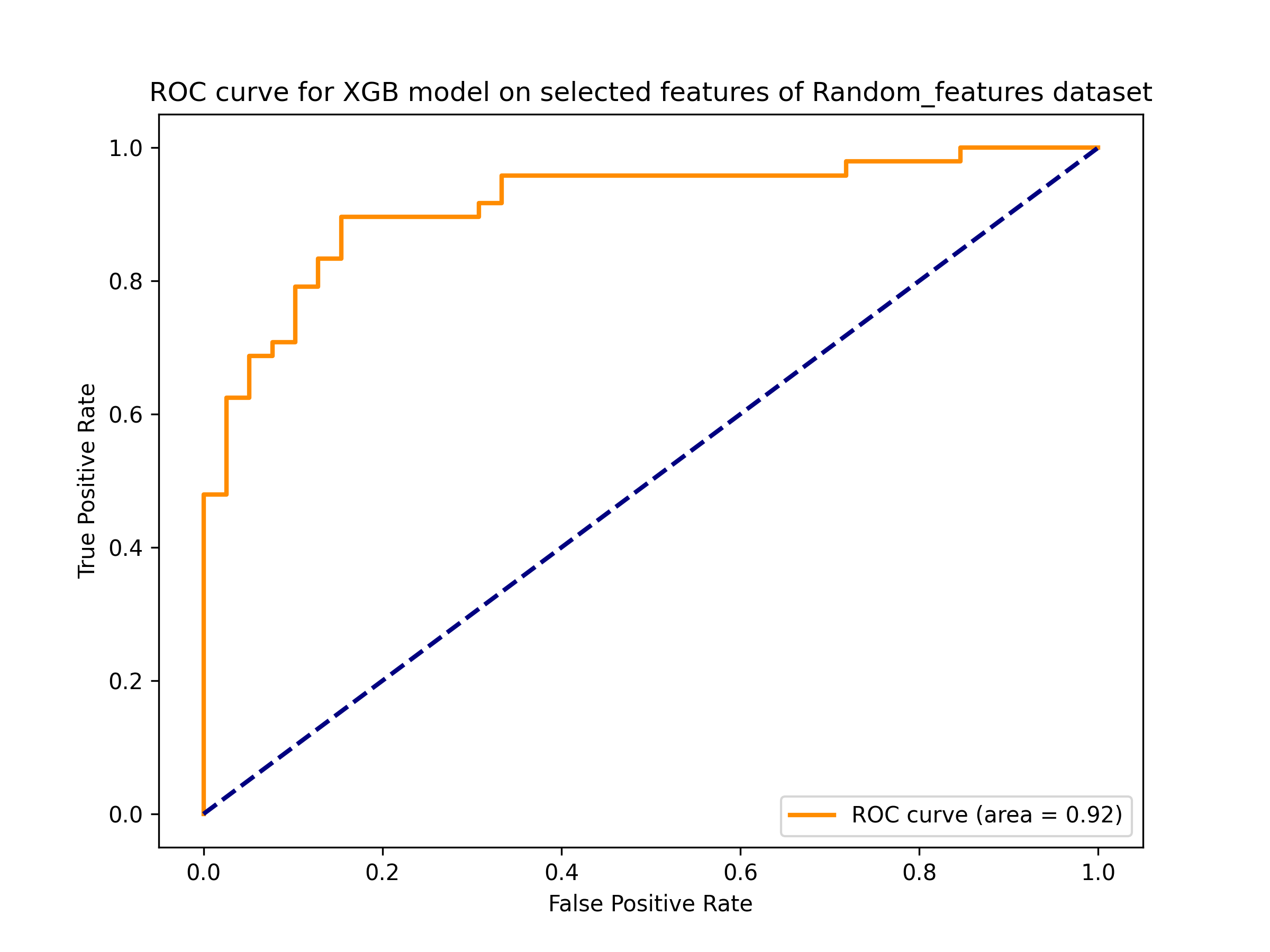
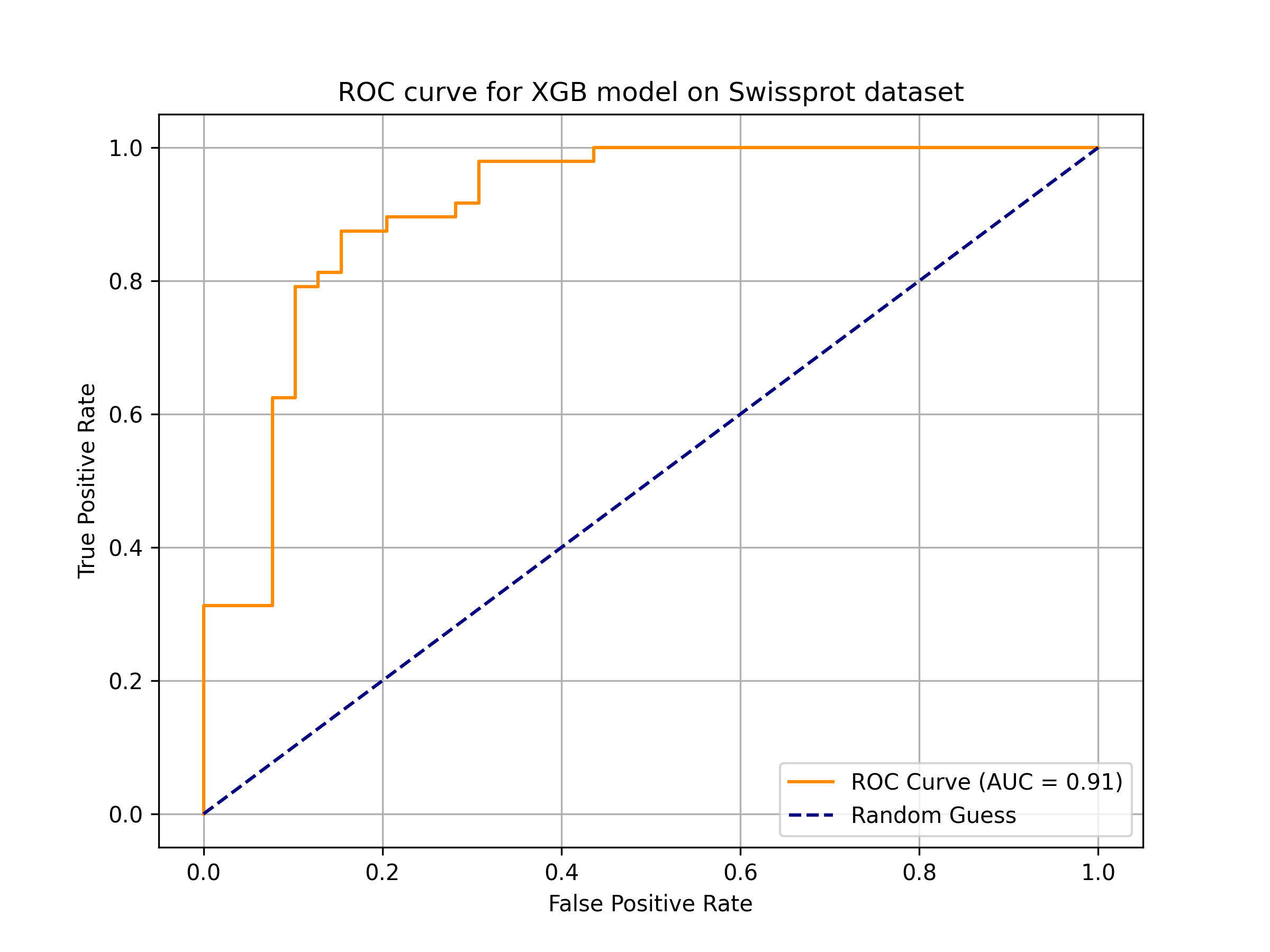
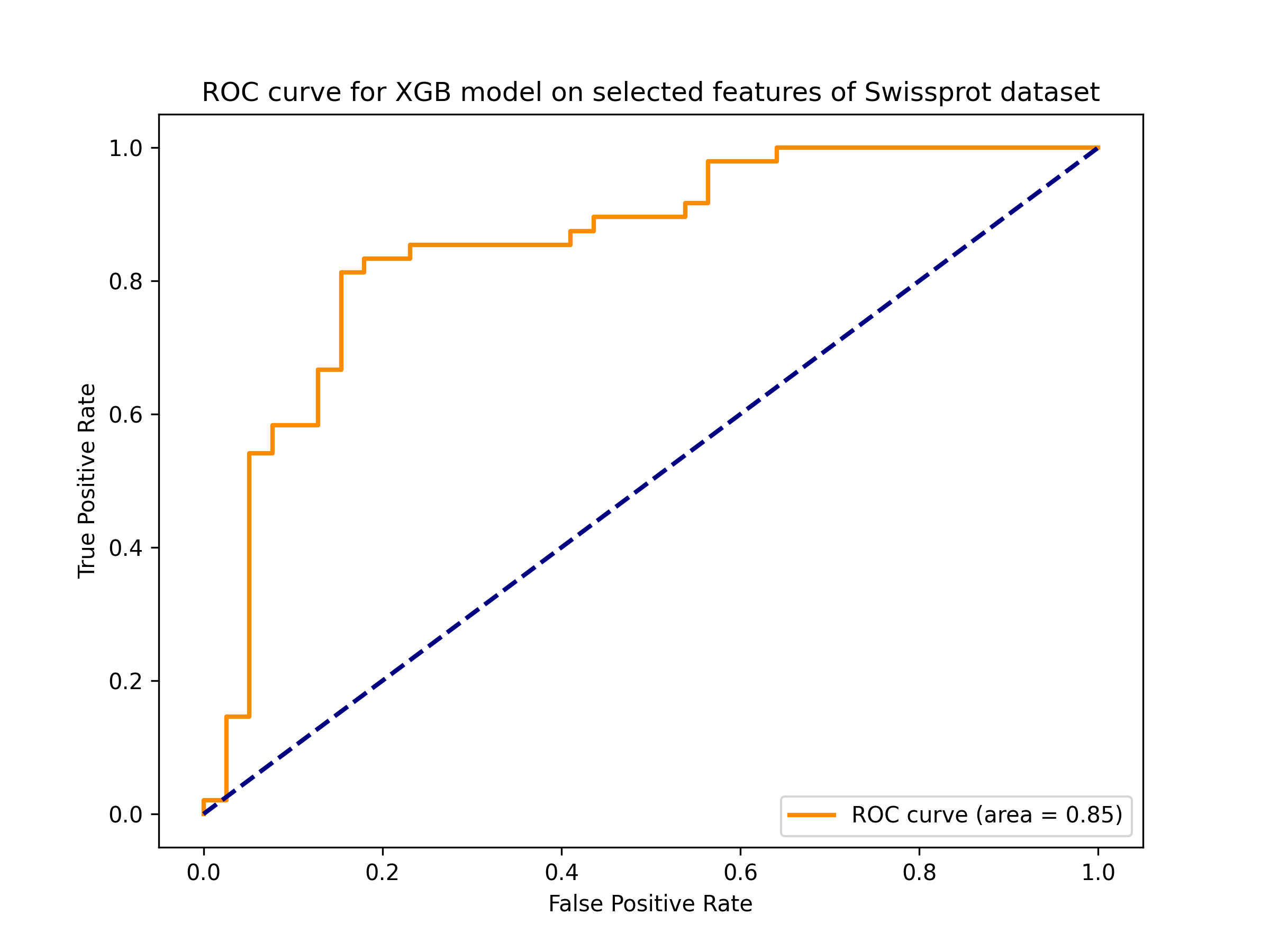
AagingPred is a machine learning-based tool designed to predict anti-aging peptides. It employs models trained on descriptors that can be calculated from amino acid sequences. The tool is capable of accepting peptide sequences in Fasta format, with lengths ranging from 5 to 50 amino acid residues.
For detailed instructions on how to utilize the tool, users are advised to refer to the Help page, which provides comprehensive information on the tool's usage, features, and functionalities.
To gain a deeper understanding of the methods and models employed by AagingPred, individuals can visit the Model page. This resource offers insights into the underlying techniques, algorithms, and data employed in the development of AagingPred's predictive models.